2025-02-12 graphs lecture 7
[[lecture-data]]
2025-02-12
1. Graph Signals and Graph Signal Processing
information theoretic threshold
Recall from last time that in the balanced sbm with
Community detection is difficult when the graph is sparse. Traditional methods may fail even before we hit this threshold!
contextual stochastic block model (C-SBM)
The (binary) contextual stochastic block model (C-SBM) is like an SBM, but includes node features that are drawn from a Gaussian distribution. Here,
- The adjacency matrix is given as
where
(or ) and
And node featuresare drawn , ,
So
Feature-aware spectral embeddings
Recall that is spectral clustering, which is fully unsupervised. For spectral embedding, we make use of some of the known community assignments.
Feature-aware spectral embeddings incorporate the availability of node features into our predictions (for example, in a C-SBM) when using spectral embedding.
Let
- Diagonalize
as - Pick the top
eigenvectors to create - Define
where are the top eigenvectors of .
Before, we had
Now, our hypothesis class is instead:
In the presence of node features, the information theoretic threshold for community detection becomes (with
the community detection is possible in an information theoretic sense when
Computational thresholds for spectral clustering (non-rigorous)
maybe there exists an algorithm that can find these communities, but can we do so in polynomial time? Can we do even better?
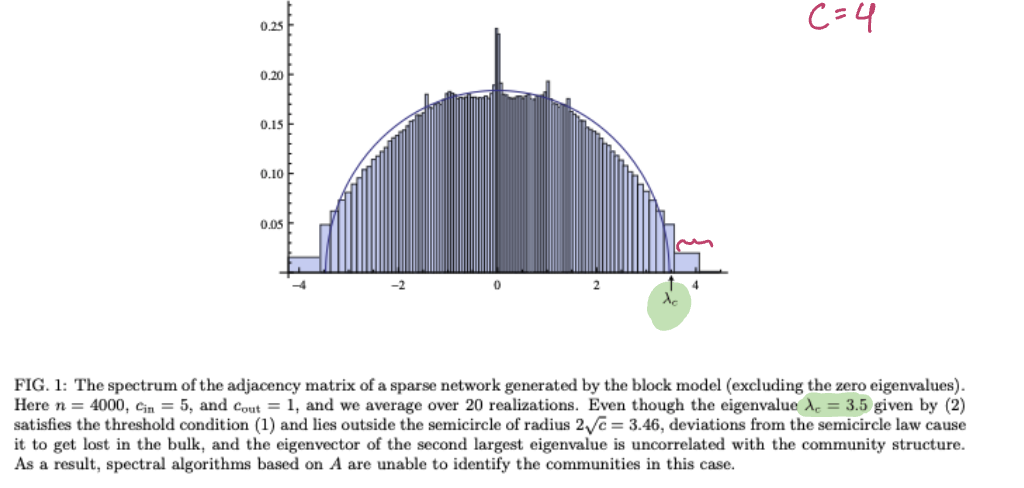
spectral redemption - Krzkala et al 2013. Computational threshold using adjacency matrix
Even by doing spectral embedding or feature-aware spectral embeddings, since we have to fix
In practice, spectral algorithms fail above the information theoretic threshold (sometimes, significantly above that threshold!)
Can GNNs help?
Let's go back to Lecture 3. We saw that the expressivity of graph convolutions can find regression solutions in many cases
Recall the conditions for finding a convolutional graph filter:
In the example above, having access to
This motivates using GNNs for semi-supervised community detection on graphs
We can use GNNs to solve the semi-supervised community detection node-level task on graphs.
Let
As long as
and for all
then there exists a graph convolution (or a 1-layer linear GNN) that approximates
where
And we can choose our hypothesis class so that
(see we can use GNNs to solve feature-aware semi-supervised learning problems)
- Homework due 2 weeks from today (first week of)
A note on sparse matrix-vector multiplications
- the usual
matmuloperation in PyTorch requiresto compute for
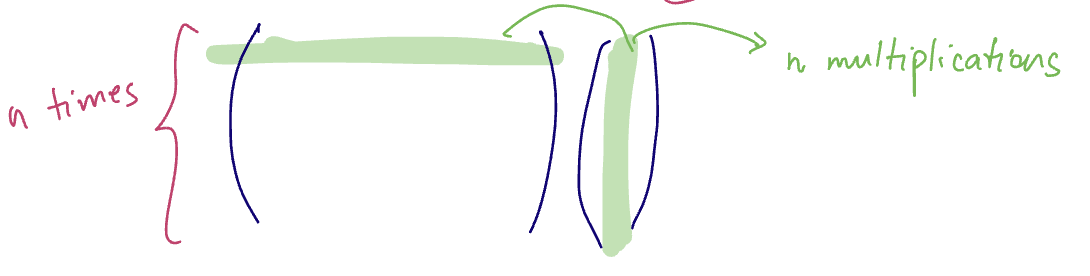
Illustration to see the
Typically the graph shift operator
- in this case,
nonzero entries can achieve complexity. But, to achieve it you need to use the sparse tensor representation of
How sparse matmuls work
Suppose we have a sparse adjacency matrix
There are two main options to store this more efficiently.
This can be better from a memory perspective, but can still be quite large if we have a few nodes with high degree. A nice solution to this is to use compressed sparse row representation instead.
(see coordinate representation)
can become
Another method to compress a sparse matrix's information is through compressed sparse row (CSR) representation - this is most popular.
Suppose
Instead of writing out the row index for each element, we can collect the column indices for each row, put each of these collections together, and then have a pointer tell us where to start reading.
If
- the
columntensor containsentries. Each entry contains the column index for one of the non-zero elements, sorted by ascending row index. roworpointertensor containsentries - the first
entries contain the starting index for where the row's elements start in the columntensor - the last entry is
- the first
valuetensor containsentries and contains the non-zero elements sorted by row then column index.
Pointer = [0 2 4 6]
Column = [0 2 2 3 0 1 1 3]
Value = [12 16 19 14 26 19 14 7]
Matrix multiplication can then become much more efficient computationally. Using sparse matrix multiplication we get
in PyTorch, to transform to a sparse tensor:
S.to_sparse()for COOS.to_sparse_csr()for CSR
matrix-vector multiplication is as before: S @ x or torch.mv(S,x) for both representations and this will give a faster multiplication.